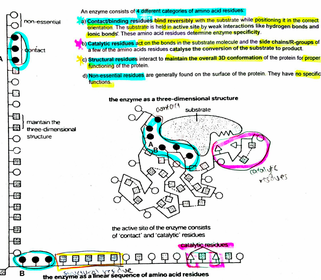
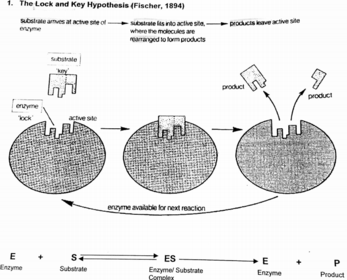
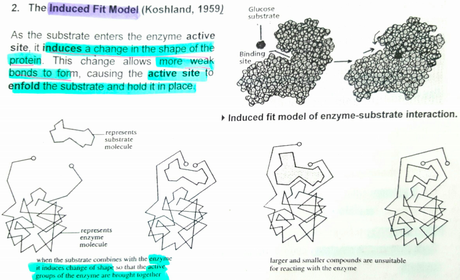
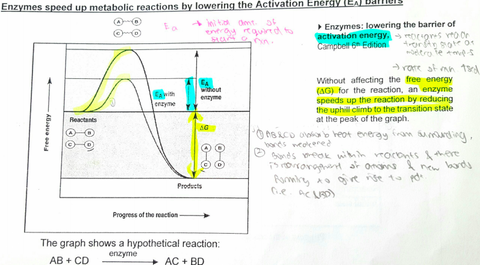
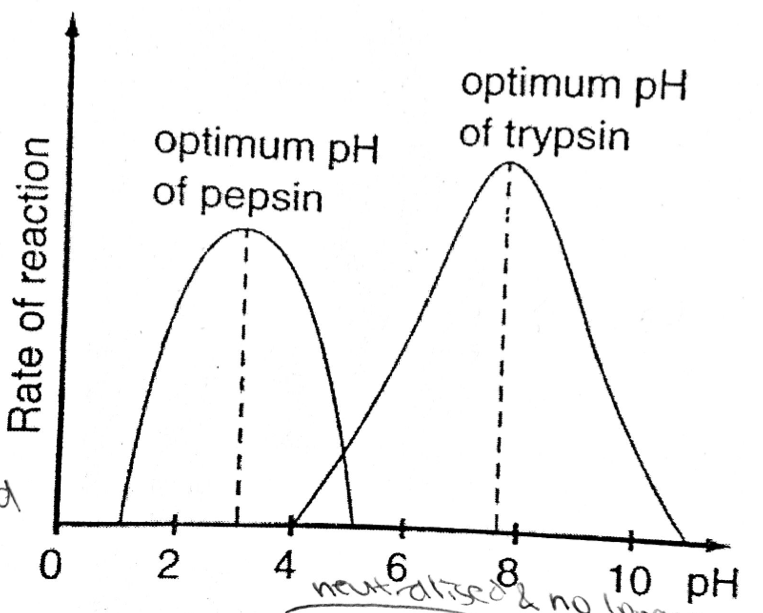
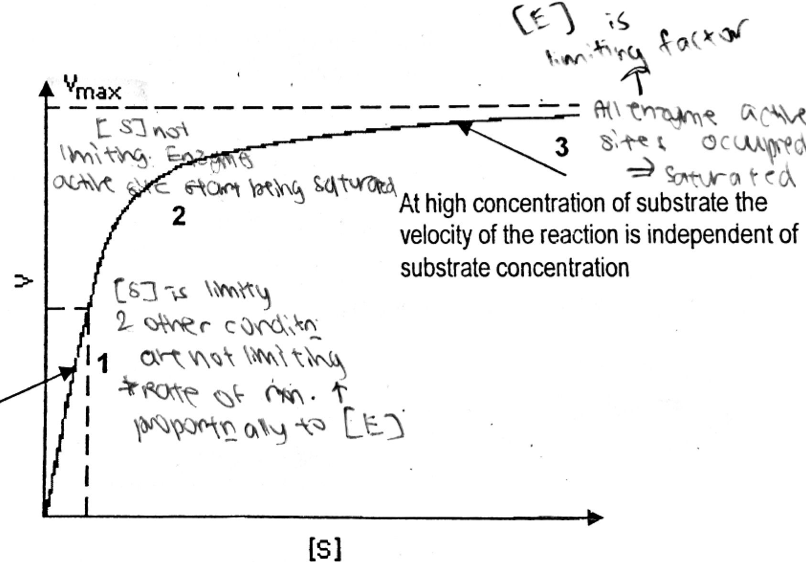
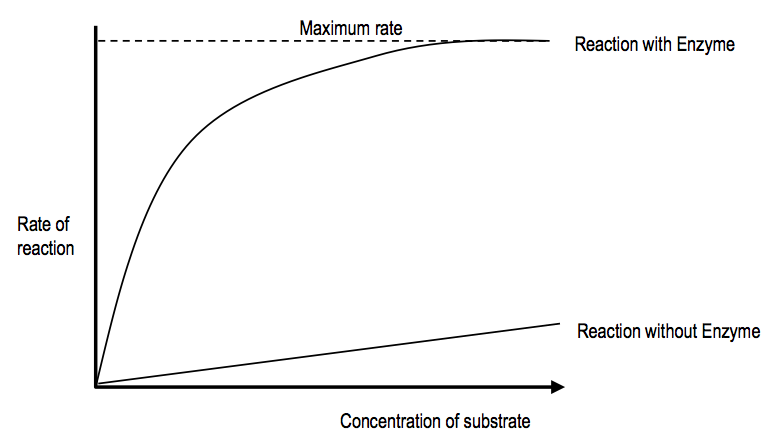
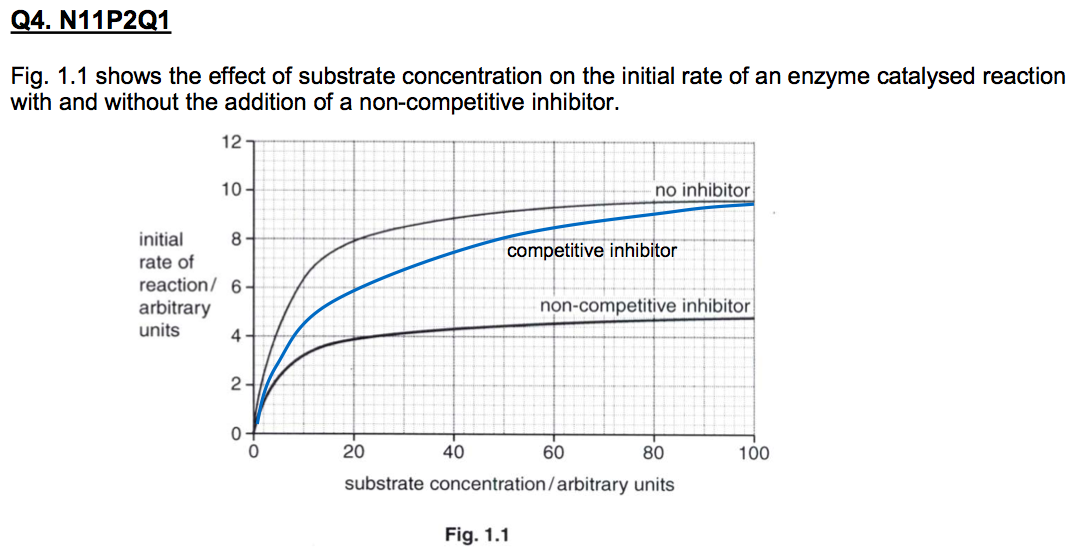
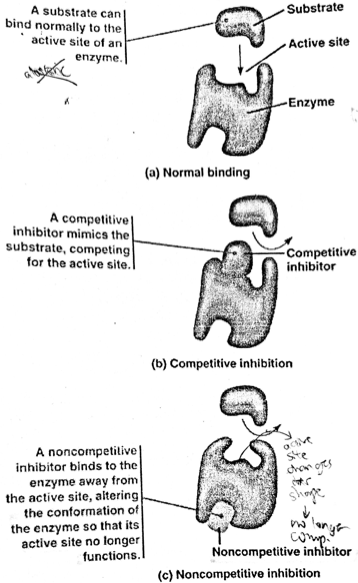
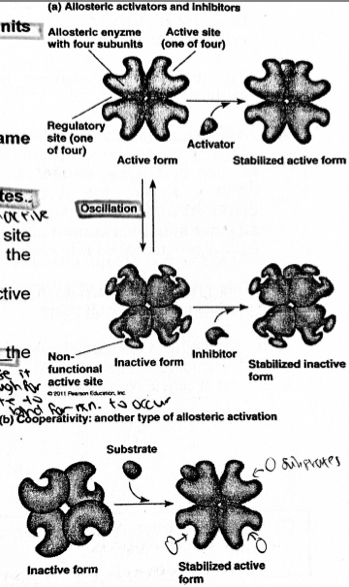
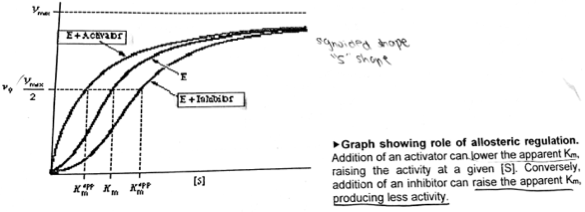
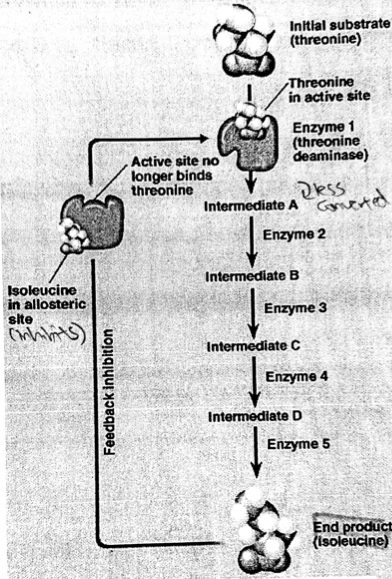
A level BIOLOGY (BIO CORE IDEA 1: The Cell & Biomolecules of Life) Karteikarten am BIO: Chapter 4 - Enzymes, erstellt von ws_ 149 am 26/03/2017.
Angeheftet an
120
1
0
Keine Merkmale angegeben
|
|
Erstellt von ws_ 149
vor mehr als 7 Jahre
|
|
Schließen